The goal of cvwrapr is to make cross-validation (CV) easy. The main function in the package is kfoldcv. It performs K-fold CV for a hyperparameter, returning the CV error for a path of hyperparameter values along with other useful information. The computeError function allows the user to compute the CV error for a range of loss functions from a matrix of out-of-fold predictions.
See the package website for more examples and documentation.
Installation
You can install cvwrapr in the following ways:
# from CRAN
install.packages("cvwrapr")
# development version from GitHub
# install.packages("devtools")
devtools::install_github("kjytay/cvwrapr")Example
This is a basic example showing how to perform cross-validation for the lambda parameter in the lasso (Tibshirani 1996).
# simulate data
set.seed(1)
nobs <- 100; nvars <- 10
x <- matrix(rnorm(nobs * nvars), nrow = nobs)
y <- rowSums(x[, 1:2]) + rnorm(nobs)
library(cvwrapr)
library(glmnet)
set.seed(1)
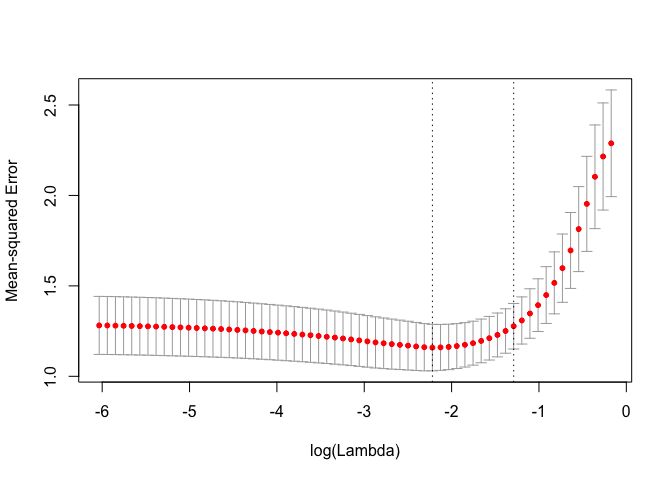
cv_fit <- kfoldcv(x, y, train_fun = glmnet, predict_fun = predict)The returned output contains information on the CV procedure and can be plotted.
names(cv_fit)
#> [1] "lambda" "cvm" "cvsd" "cvup" "cvlo"
#> [6] "lambda.min" "lambda.1se" "index" "name" "overallfit"
plot(cv_fit)